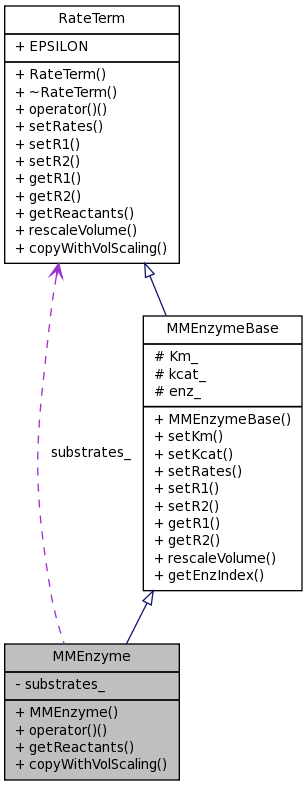
MMEnzyme Class Reference
#include <RateTerm.h>
Public Member Functions | |
| MMEnzyme (double Km, double kcat, unsigned int enz, RateTerm *sub) | |
| double | operator() (const double *S) const |
| Computes the rate. The argument is the molecule array. | |
| unsigned int | getReactants (vector< unsigned int > &molIndex) const |
| RateTerm * | copyWithVolScaling (double vol, double sub, double prd) const |
Constructor & Destructor Documentation
| MMEnzyme::MMEnzyme | ( | double | Km, | |
| double | kcat, | |||
| unsigned int | enz, | |||
| RateTerm * | sub | |||
| ) | [inline] |
Referenced by copyWithVolScaling().
Member Function Documentation
| RateTerm* MMEnzyme::copyWithVolScaling | ( | double | vol, | |
| double | sub, | |||
| double | prd | |||
| ) | const [inline, virtual] |
Duplicates rate term and then applies volume scaling. Arguments are volume of reference voxel, product of vol/refVol for all substrates: applied to R1 product of vol/refVol for all products: applied to R2
Note that unless the reaction is cross-compartment, the vol/refVol will be one.
Implements RateTerm.
References MMEnzymeBase::enz_, MMEnzymeBase::kcat_, MMEnzymeBase::Km_, MMEnzyme(), and NA.

| unsigned int MMEnzyme::getReactants | ( | vector< unsigned int > & | molIndex | ) | const [inline, virtual] |
This function finds the reactant indices in the vector S. It returns the number of substrates found, which are the first entries in molIndex. The products are the remaining ones. Note that it does NOT find products for unidirectional reactions, which is a bit of a problem.
Implements RateTerm.
References MMEnzymeBase::enz_, and RateTerm::getReactants().

| double MMEnzyme::operator() | ( | const double * | S | ) | const [inline, virtual] |
Computes the rate. The argument is the molecule array.
Implements RateTerm.
References MMEnzymeBase::enz_, RateTerm::EPSILON, MMEnzymeBase::kcat_, and MMEnzymeBase::Km_.
The documentation for this class was generated from the following file:
- ksolve/RateTerm.h
 1.6.1
1.6.1